Nucleic Acid Sequence-Based Amplification, a New Method for Analysis of Spliced and Unspliced Epstein-Barr Virus Latent Transcripts, and Its Comparison with Reverse Transcriptase PCR
Por um escritor misterioso
Descrição

Epstein–Barr virus co-opts TFIIH component XPB to specifically
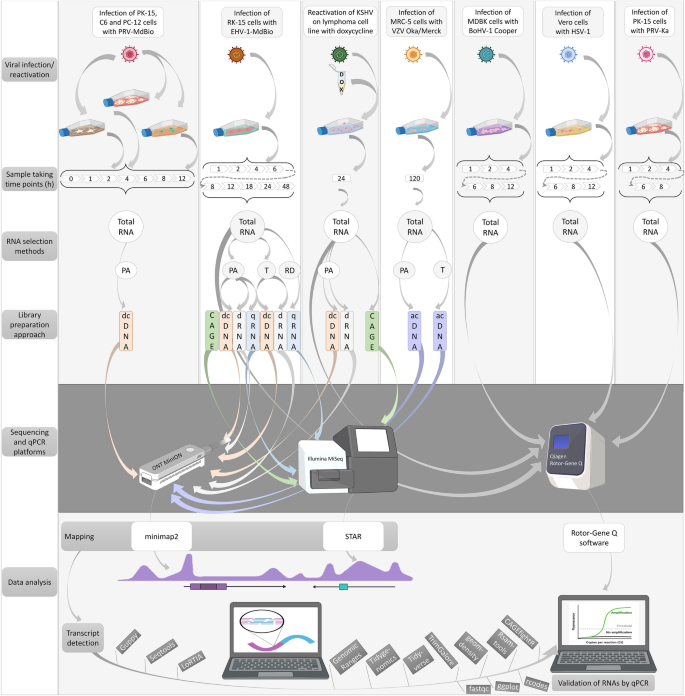
Identification of herpesvirus transcripts from genomic regions

RNA-based isothermal amplification technology and its clinical

BHRF1, the Epstein-Barr Virus (EBV) Homologue of the BCL-2 Proto

PDF) Nucleic acid sequence based amplification (NASBA) - prospects

DNA nucleic acid sequence-based amplification-based genotyping for
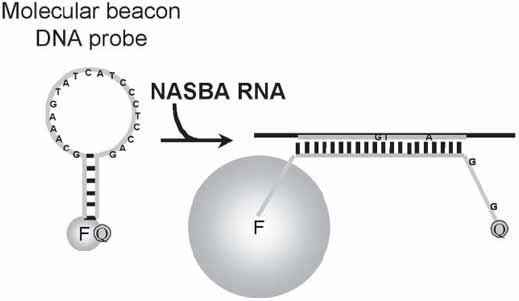
Nucleic Acid Sequence-Based Amplification
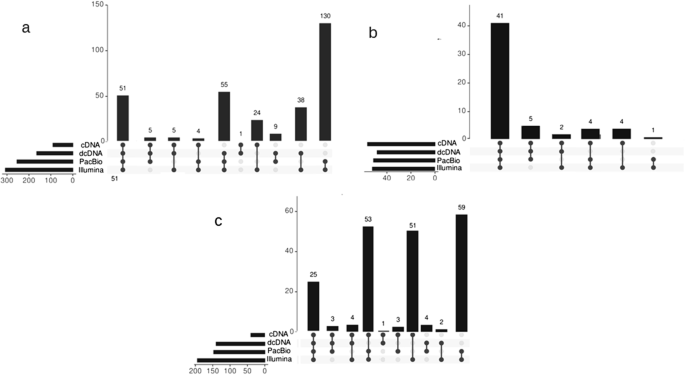
Integrative profiling of Epstein–Barr virus transcriptome using a

BamHI‐A rightward frame 1, an Epstein–Barr virus‐encoded oncogene
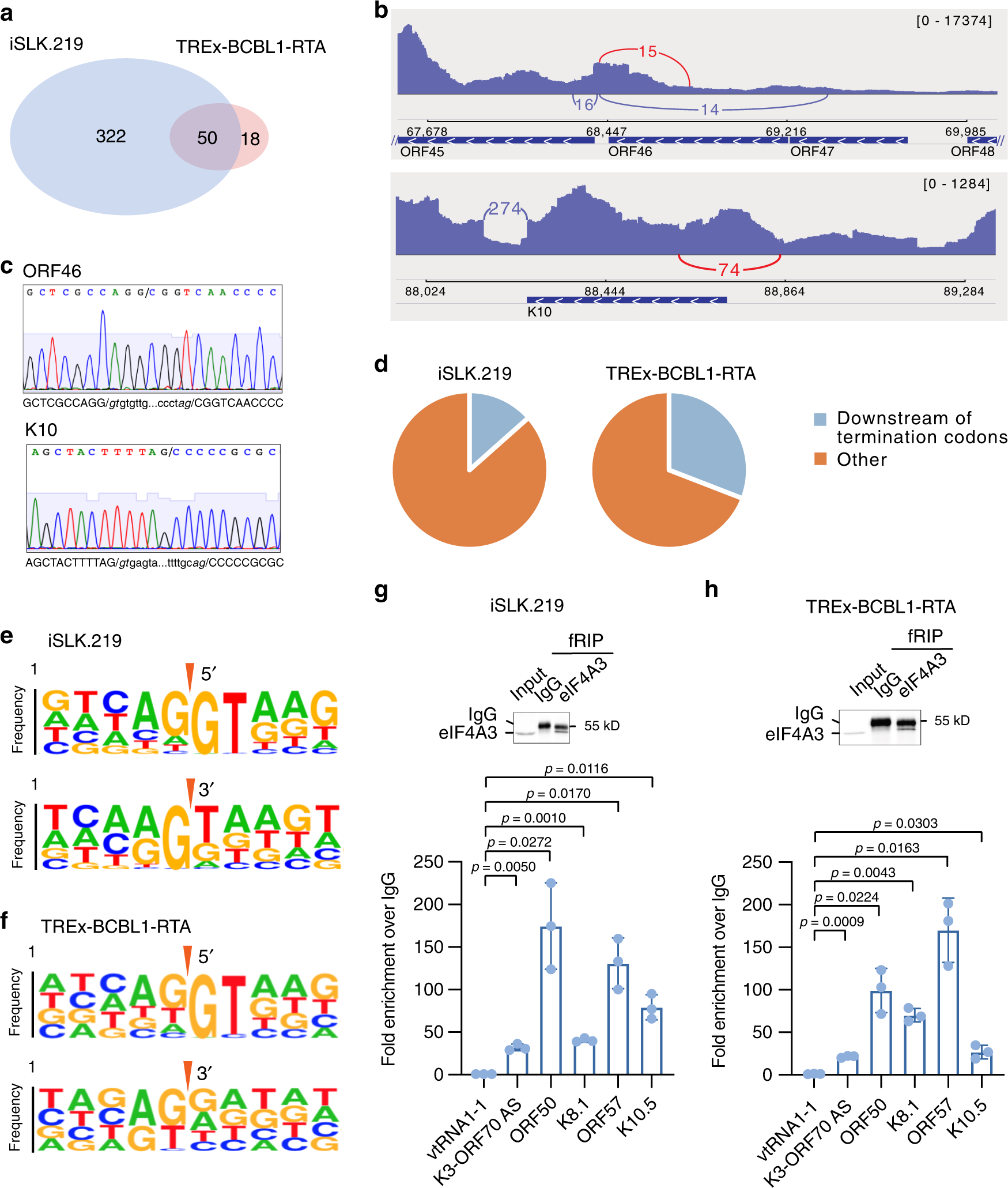
The RNA quality control pathway nonsense-mediated mRNA decay
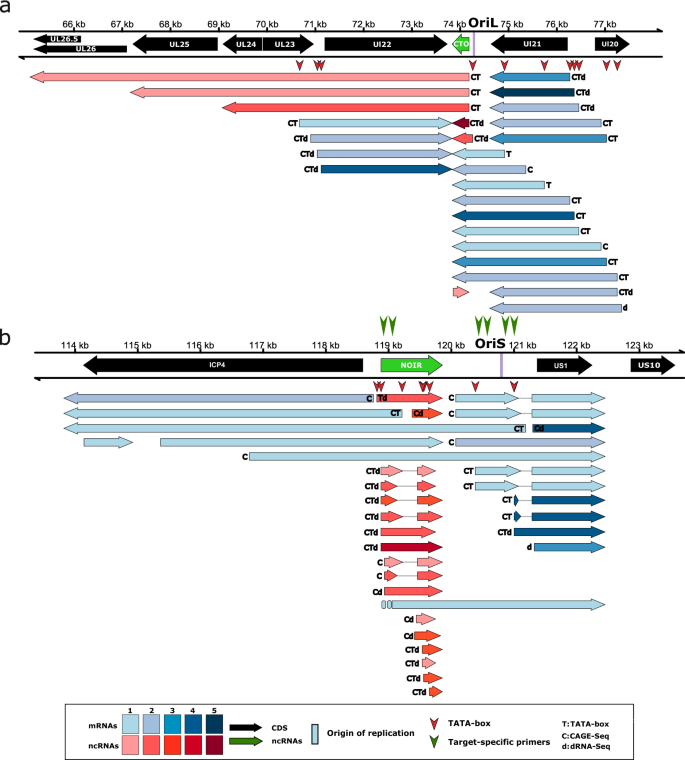
Identification of herpesvirus transcripts from genomic regions

Intron Retention May Regulate Expression of Epstein-Barr Virus

Nucleic Acid Sequence Based Amplification - an overview

Rapid and effective detection of Macrobrachium rosenbergii
de
por adulto (o preço varia de acordo com o tamanho do grupo)






