Dynamic allocation of orthogonal ribosomes facilitates uncoupling
Por um escritor misterioso
Descrição

Team:Rice/Description

Team:ShanghaiTech/Project Oribosome

Modelling co-translational dimerization for programmable nonlinearity in synthetic biology
Operation of the negative feedback controller. a. Structure and
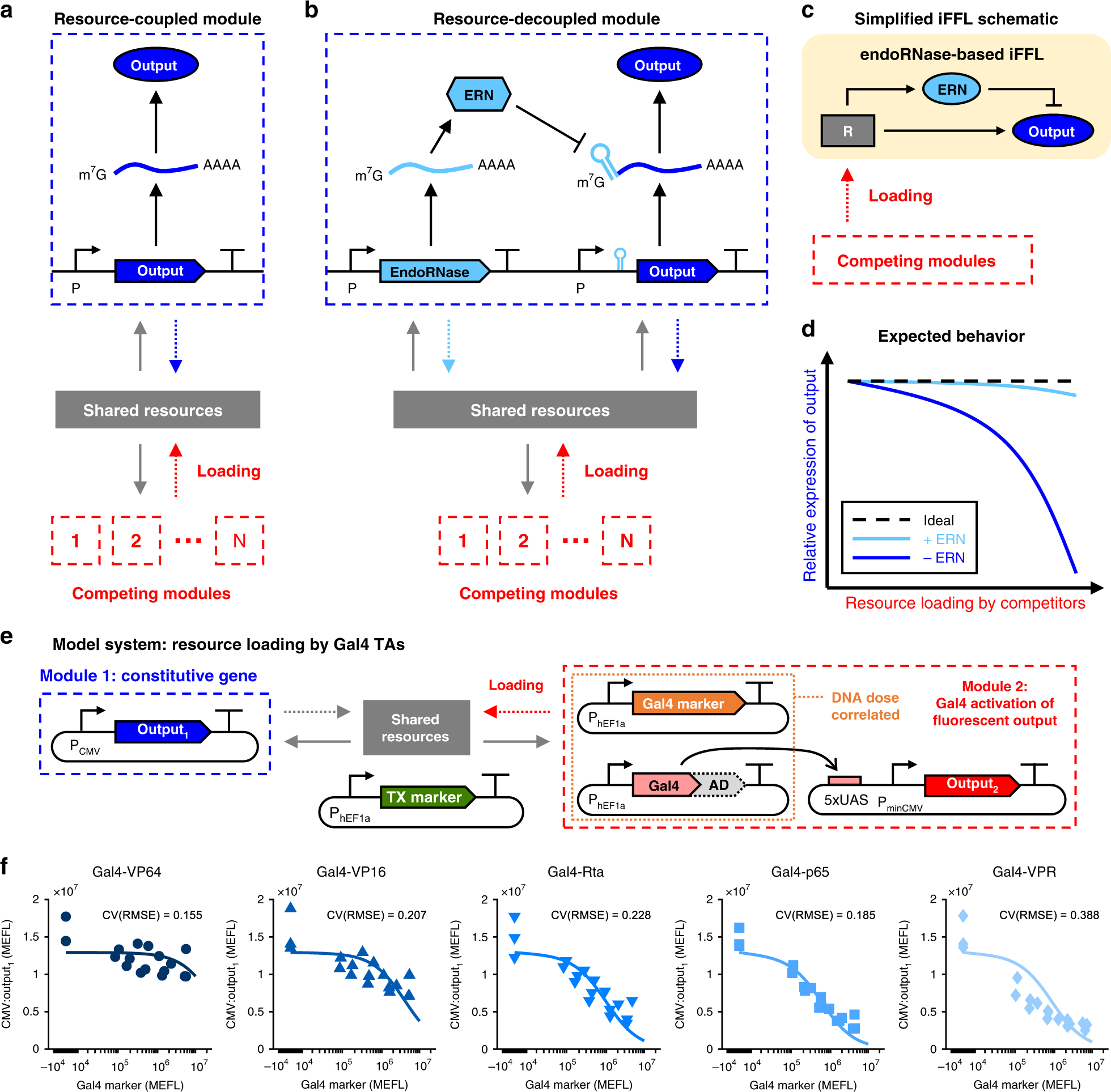
An endoribonuclease-based feedforward controller for decoupling resource-limited genetic modules in mammalian cells

The interplay between growth rate and nutrient quality defines gene expression capacity

Synthetic Biology

Orthogonal translation enables heterologous ribosome engineering in E. coli. - Abstract - Europe PMC

PDF] A Minimal Model of Ribosome Allocation Dynamics Captures Trade-offs in Expression between Endogenous and Synthetic Genes.
GitHub - apsduk/Nat-Commun-2018: MATLAB code for our paper Dynamic allocation of orthogonal ribosomes facilitates uncoupling of co-expressed genes

Synthetic Biology Breakthrough Means Bacteria Could Produce Our Essential Drugs

PDF] Isocost Lines Describe the Cellular Economy of Genetic Circuits
de
por adulto (o preço varia de acordo com o tamanho do grupo)